projected_graph¶
- projected_graph(B, nodes, multigraph=False)[source]¶
Returns the projection of B onto one of its node sets.
Returns the graph G that is the projection of the bipartite graph B onto the specified nodes. They retain their attributes and are connected in G if they have a common neighbor in B.
Parameters : B : NetworkX graph
The input graph should be bipartite.
nodes : list or iterable
Nodes to project onto (the “bottom” nodes).
multigraph: bool (default=False) :
If True return a multigraph where the multiple edges represent multiple shared neighbors. They edge key in the multigraph is assigned to the label of the neighbor.
Returns : Graph : NetworkX graph or multigraph
A graph that is the projection onto the given nodes.
See also
is_bipartite, is_bipartite_node_set, sets, weighted_projected_graph, collaboration_weighted_projected_graph, overlap_weighted_projected_graph, generic_weighted_projected_graph
Notes
No attempt is made to verify that the input graph B is bipartite. Returns a simple graph that is the projection of the bipartite graph B onto the set of nodes given in list nodes. If multigraph=True then a multigraph is returned with an edge for every shared neighbor.
Directed graphs are allowed as input. The output will also then be a directed graph with edges if there is a directed path between the nodes.
The graph and node properties are (shallow) copied to the projected graph.
Examples
>>> from networkx.algorithms import bipartite >>> B = nx.path_graph(4) >>> G = bipartite.projected_graph(B, [1,3]) >>> print(G.nodes()) [1, 3] >>> print(G.edges()) [(1, 3)]
If nodes
 , and
, and 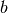 are connected through both nodes 1 and 2 then
building a multigraph results in two edges in the projection onto
[
are connected through both nodes 1 and 2 then
building a multigraph results in two edges in the projection onto
[ ,`b`]:
,`b`]:>>> B = nx.Graph() >>> B.add_edges_from([('a', 1), ('b', 1), ('a', 2), ('b', 2)]) >>> G = bipartite.projected_graph(B, ['a', 'b'], multigraph=True) >>> print([sorted((u,v)) for u,v in G.edges()]) [['a', 'b'], ['a', 'b']]